About ALLOY
ALLOY is a fast and accurate method for local ancestry inference, which is the problem of labeling each position of the genome with an ancestral origin.
It uses a factorial HMM that incorporates a flexible background model of linkage disequilibrium in ancestral populations. Previous methods used either simplified models of LD or large LD models that explicitly model ancestral haplotypes (e.g. AncestryMap, LAMP, WINPOP, SABER, HAPMIX, and HAPAA)
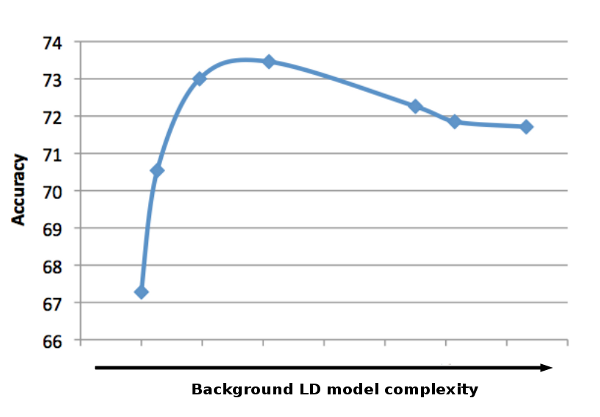
What makes ALLOY unique is that it can efficiently incorporate a variety of background LD models of intermediate complexity, resulting in better performance than models that are too simple or too large. Further description of the explored models and their relationship to the models of previous work are detailed in the paper.
Download
ALLOY is written in Java, and is available here: alloy_v0.2.zip
Publication
Ancestry inference in complex admixtures via variable-length Markov chain linkage models.
Journal of Computational Biology,
20.3 (2013): 199-211.
Ancestry inference in complex admixtures via variable-length Markov chain linkage models.
Proceedings of the Sixteenth Annual Conference on Research in Computational Molecular Biology (RECOMB 2012),
pp 12-28.
Questions?
Contact Jesse Rodriguez: jesserod at cs dot stanford dot edu